Kehui "Coffee" Xiang
A passionate scientist who studies gene regulation. I combine high-throughput system methodologies, RNA and protein biochemistry, functional genomic screens, and machine learning to unravel gene-regulatory mechanisms.

I am a scientist driven by my curiosity for the unknown. I have a multidisciplinary background in mathematics, physics, and biology. I combine wet-bench labwork with computational modeling to address the most exciting biological and biomedical questions. I am currently working as a Postdoc Associate with Dr. David Bartel at the Whitehead Institute.
Presently, I am working on
Using machine learning to identify genetic variants associated with human reproductive disorders;
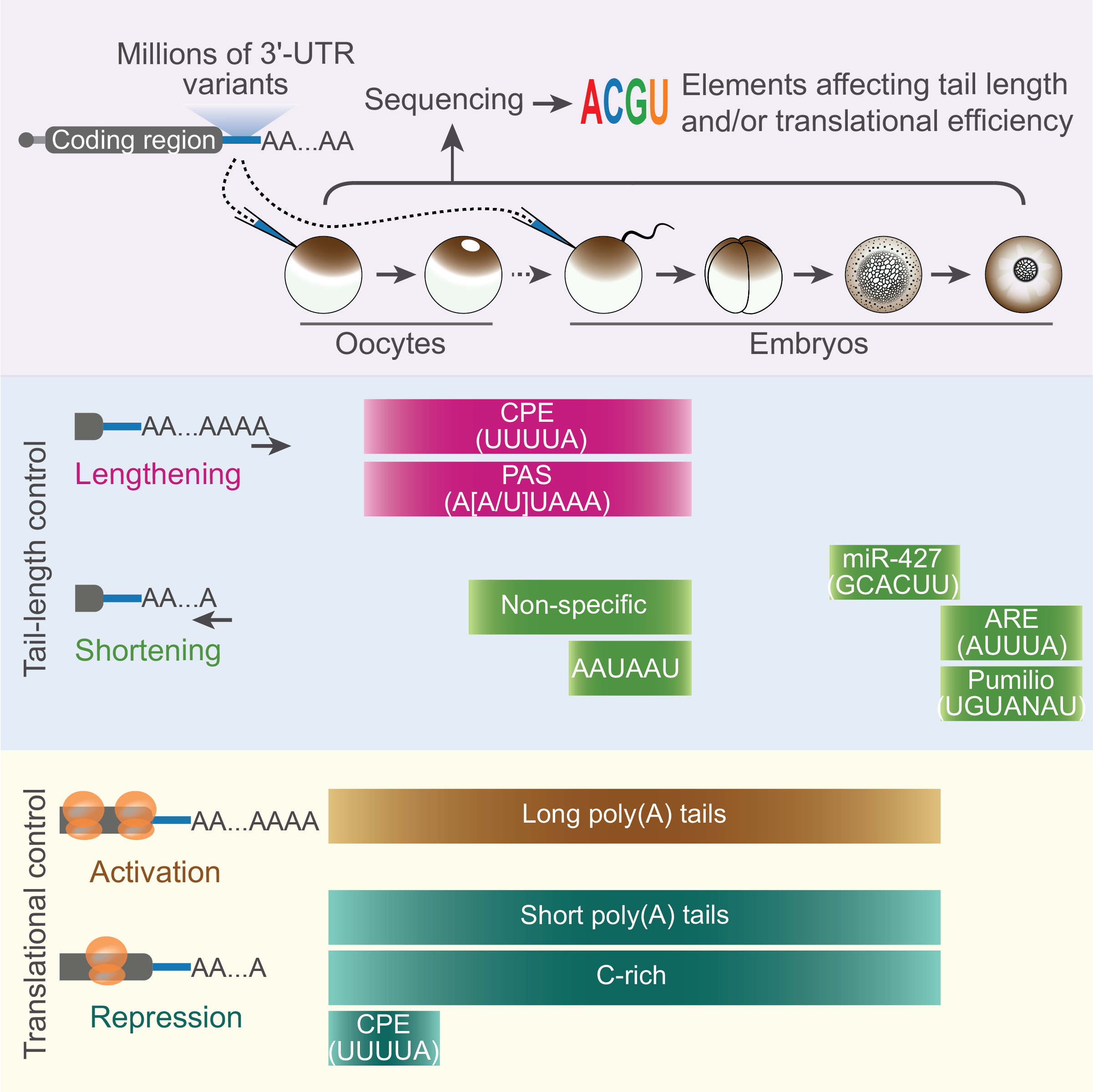
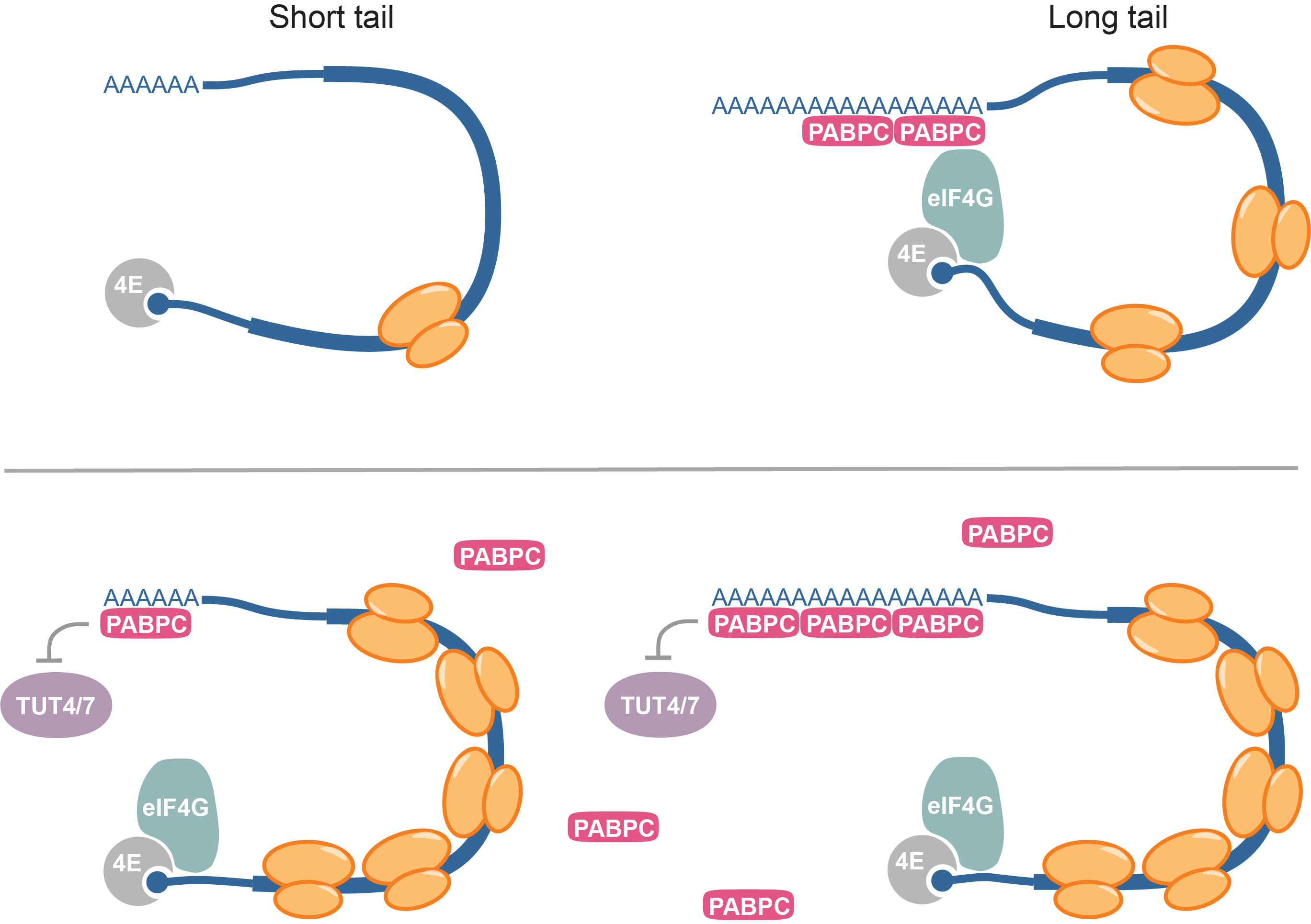
Post-transcriptional gene regulation during veterbrate early development;
Novel technologies to study translational control mechanisms in mammalian cells.
Outside my work, I like building things, exploring the nature, making videos, exercises, games, and movies.
I worked on multiple projects about post-transcriptional gene regulation.
I carried out structural and biochemical studies on transcription-coupled mRNA 3’-end processing.